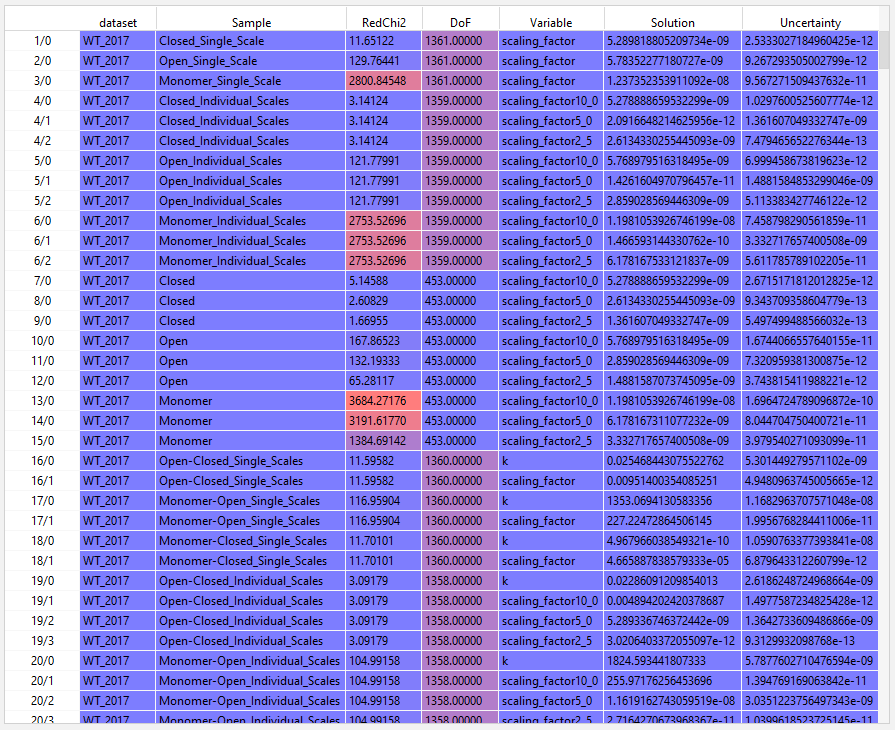
Here is an example demonstrating some of the logic that you wrote for concentrations and errors, with Pandas:
import numpy as np import pandas as pd MAX_VARS = 20 MW = 42_000 def load(filename: str = 'scaled_uncertanty_fits.txt') -> pd.DataFrame: df = pd.read_csv( filename, sep=',|±', skiprows=1, # skip the original headers - they aren't wide enough names=[ 'Sample', 'RedChi2', 'DoF', *range(3*MAX_VARS), ] ) df.index.name = 'csv_index' # Forward-fill the dataset name (e.g. WT_2017) is_dataset = df.iloc[:, 1].isna() df.insert(loc=0, column='dataset', value=df.loc[is_dataset, 'Sample']) df['dataset'].ffill(inplace=True) return df[~is_dataset] def normalize_vars(df: pd.DataFrame) -> pd.DataFrame: """Normalize variable-solution-uncertainty triples""" var_offset = 4 var_cols = df.iloc[:, var_offset:] meta_names = pd.Index(name='varmeta', data=['Variable', 'Solution', 'Uncertainty']) rectangular = pd.DataFrame( index=df.index, columns=pd.MultiIndex.from_product(( meta_names, pd.RangeIndex(name='varno', start=0, stop=MAX_VARS), )) ) n_vars = var_cols.notna().sum(axis=1)//3 for row_vars, group in var_cols.groupby(n_vars): source = group.iloc[:, :row_vars] source.columns = pd.MultiIndex.from_product( (('Variable',), range(row_vars)), names=('varmeta', 'varno'), ) rectangular.loc[group.index, source.columns] = source source = group.iloc[:, row_vars: row_vars*2] source.columns = pd.MultiIndex.from_product( (('Solution',), range(row_vars)), names=('varmeta', 'varno'), ) rectangular.loc[group.index, source.columns] = source source = group.iloc[:, row_vars*2: row_vars*3] source.columns = pd.MultiIndex.from_product( (('Uncertainty',), range(row_vars)), names=('varmeta', 'varno'), ) rectangular.loc[group.index, source.columns] = source long = rectangular.stack(level='varno') normalized = ( pd.merge(left=df[['dataset', 'Sample']], right=long[meta_names], on='csv_index') .set_index(['dataset', 'Sample', 'Variable'], append=True) .astype({'Solution': float, 'Uncertainty': float}) .unstack('Variable') ) return normalized def extract_factors(df: pd.DataFrame): names = df.columns[df.columns.get_loc('Solution')].droplevel(0) values = ( names.get_level_values('Variable') .to_series(name='factor', index=names) .str.replace('_', '.') .str.extract(r'(\d+\.\d+)$', expand=False) .dropna() .astype(float) ) return values def main() -> None: df = load() df = normalize_vars(df) # For all datasets and Sample ~ Open-Closed* open_closed = df[df.index.get_level_values('Sample').str.contains('Open-Closed')] # io is converted concentrations from the number embedded in the scaling_factornnn names factors = extract_factors(df) io = conc = factors * 1e9/MW k = open_closed.loc[:, ('Solution', 'k')] k_error = open_closed.loc[:, ('Uncertainty', 'k')] fo = k/(k + 1) fc = 1/(k + 1) fm = 0 h = 1e-8 kh = k + h dk_fo = kh/(kh + 1) dk_fc = 1/(kh + 1) dk_fm = 0 open_errors = k_error**2 * dk_fo**2 closed_errors = k_error**2 * dk_fc**2 monomer_errors = k_error**2 * dk_fm**2 if __name__ == '__main__': main()