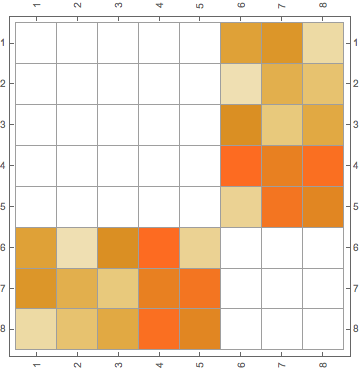
In a bipartite graphs there are two class of nodes, say A and B. Each node of class A may interact only with nodes of the class B, and viceversa. The corresponding adjecency matrix is thus composed by two off-diagonal blocks (if you sort the vertices in an appropriate way).
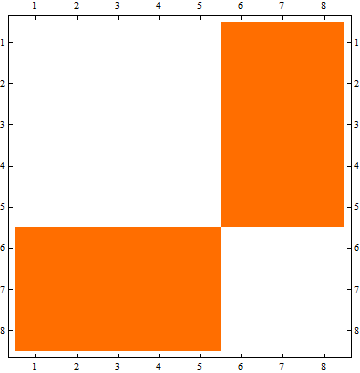
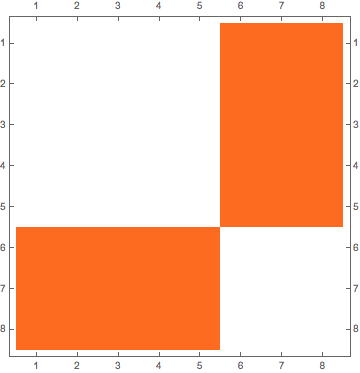
I realize that when working with adjecency matrix for a given bipartite graphs, sometimes I get the desired order in the adjecency matrix, and thus if I plot it, it is formed by two off-diagonal blocks:
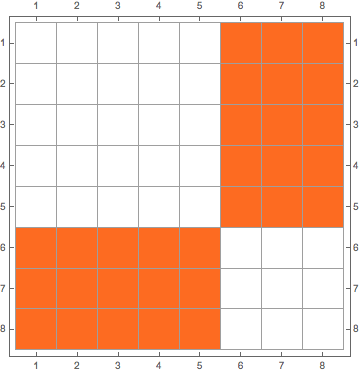
g1=CompleteGraph[{5,3}] m1=AdjacencyMatrix[g1] // MatrixPlot 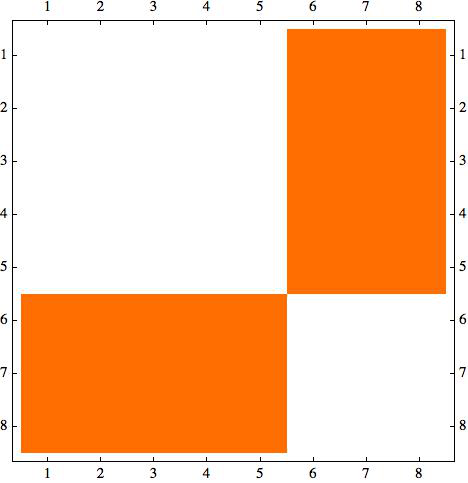
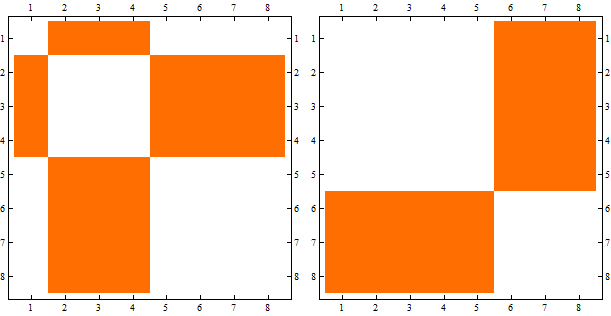
However, sometimes if I plot the adjecency matrix directly from the bipartite graphs, now it is not in the desired order:
l2 = CompleteGraph[{5, 3}] // EdgeList; weights = RandomReal[{0.5, 2}, Length[l2]]; g2 = Graph[l2, EdgeWeight -> weights, GraphLayout -> "BipartiteEmbedding"] m2=AdjacencyMatrix[g2] // MatrixPlot 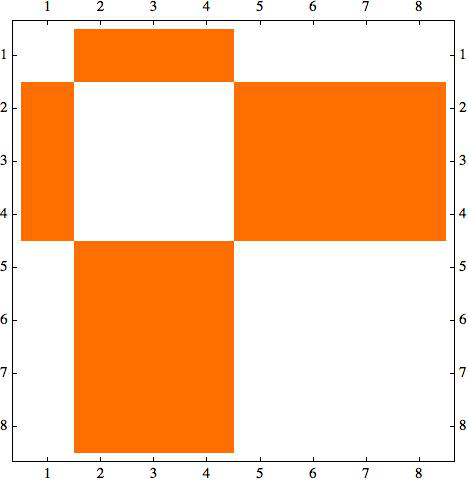
I'm trying to find a way to automatically sort m2 so to tranform it in a two off-diagonal blocks matrix.