This is a plot of my data
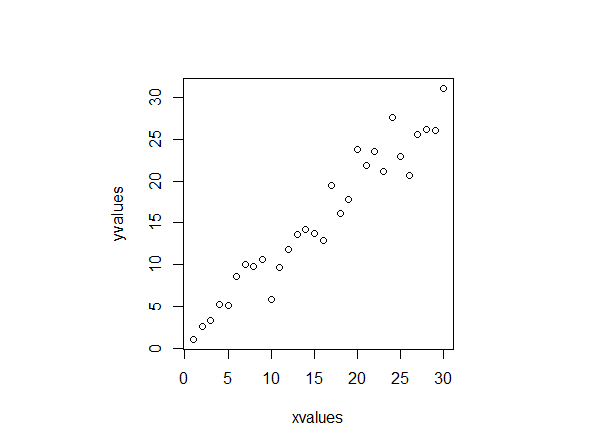
These are the values:
xvalues yvalues 1 1.091186 2 2.653722 3 3.309146 4 5.206479 5 5.115582 6 8.537005 7 10.013147 8 9.802291 9 10.667769 10 5.809750 11 9.624475 12 11.806013 13 13.587066 14 14.146781 15 13.707472 16 12.891355 17 19.435301 18 16.122108 19 17.768536 20 23.813027 21 21.819081 22 23.556074 23 21.170983 24 27.621148 25 22.932580 26 20.704689 27 25.530339 28 26.227371 29 26.051016 30 31.047145 I now do a PCA and a biplot of it:
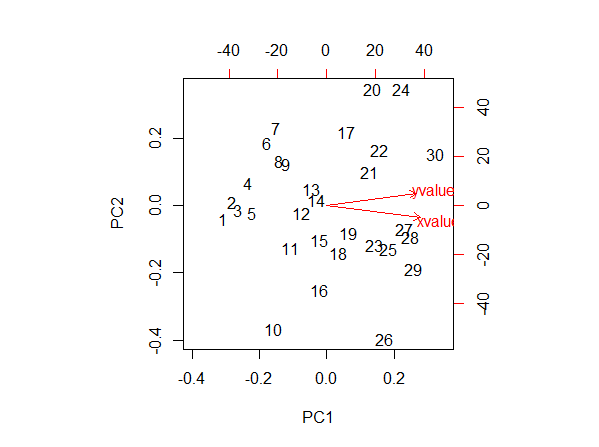
According to Jeromy Anglim in: Interpretation of biplots in principal components analysis in R
The left and bottom axes are showing the loadings; the top and right axes are showing principal component scores.The left and bottom axes are showing [normalized] principal component scores; the top and right axes are showing the loadings.
I want to be sure I really get this.
Let's start with the loadings: these can be visualized in R by writing:
results <- prcomp(your_data) results$rotation PC1 PC2 xvalues 0.7235616 -0.6902599 yvalues 0.6902599 0.7235616 summary(results) Importance of components: PC1 PC2 Standard deviation 12.0747 1.56606 Proportion of Variance 0.9835 0.01654 Cumulative Proportion 0.9835 1.00000 Now lets look at the red arrow of xvalues. Its tip is around 0.25 in the x-axis of the loadings. But according to the loadings I have just writen, it should be around 0.72. What am I missing?
Finally, lets look at the point 1. According to the axes, that is telling me the principal component score. Is it that the coordinate in the new frame of reference? It doesn't make sense to me because I think that the new origin of the axes is around the point (15,15) in the Plot 3. If I look at that (and I guess that I am completely wrong here), the point one should have a coordinate around -20 or so, and not 40. Where is my mistake?
Update
I tried plotting this:
plot(pca_results$x) 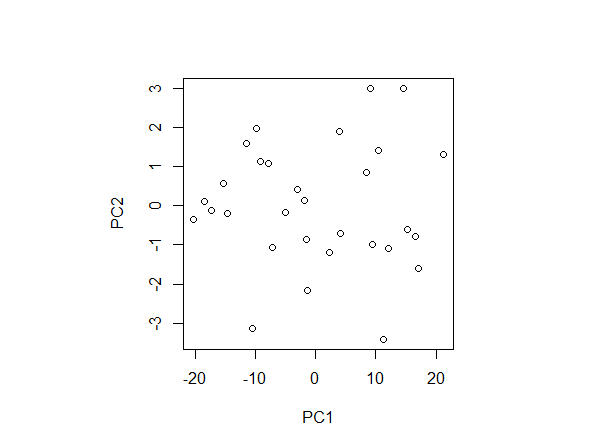
Here it can be seen that the first point has the coordinate that I thought it had to have. But, still, what are the units in the biplot then??

results$rotationfigures you present are clearly the eigenvector values, not the loadings. Please be aware that theRPCA package you use misuses the word "loadings", incorrectly calling eigenvectors "loadings". $\endgroup$results$rotationmatrix actually is. It could be eigenvectors (prior rotation) or it could the orthogonal rotation matrix. Because you didn't show the original data (the values) and full output of your PCA, I can't say. $\endgroup$