Using a biplot of values obtained through principal component analysis, it is possible to explore the explanatory variables that make up each principle component. Is this also possible with Linear Discriminant Analysis?
Examples provided use the The data is "Edgar Anderson's Iris Data" (http://en.wikipedia.org/wiki/Iris_flower_data_set). Here is the iris data:
id SLength SWidth PLength PWidth species 1 5.1 3.5 1.4 .2 setosa 2 4.9 3.0 1.4 .2 setosa 3 4.7 3.2 1.3 .2 setosa 4 4.6 3.1 1.5 .2 setosa 5 5.0 3.6 1.4 .2 setosa 6 5.4 3.9 1.7 .4 setosa 7 4.6 3.4 1.4 .3 setosa 8 5.0 3.4 1.5 .2 setosa 9 4.4 2.9 1.4 .2 setosa 10 4.9 3.1 1.5 .1 setosa 11 5.4 3.7 1.5 .2 setosa 12 4.8 3.4 1.6 .2 setosa 13 4.8 3.0 1.4 .1 setosa 14 4.3 3.0 1.1 .1 setosa 15 5.8 4.0 1.2 .2 setosa 16 5.7 4.4 1.5 .4 setosa 17 5.4 3.9 1.3 .4 setosa 18 5.1 3.5 1.4 .3 setosa 19 5.7 3.8 1.7 .3 setosa 20 5.1 3.8 1.5 .3 setosa 21 5.4 3.4 1.7 .2 setosa 22 5.1 3.7 1.5 .4 setosa 23 4.6 3.6 1.0 .2 setosa 24 5.1 3.3 1.7 .5 setosa 25 4.8 3.4 1.9 .2 setosa 26 5.0 3.0 1.6 .2 setosa 27 5.0 3.4 1.6 .4 setosa 28 5.2 3.5 1.5 .2 setosa 29 5.2 3.4 1.4 .2 setosa 30 4.7 3.2 1.6 .2 setosa 31 4.8 3.1 1.6 .2 setosa 32 5.4 3.4 1.5 .4 setosa 33 5.2 4.1 1.5 .1 setosa 34 5.5 4.2 1.4 .2 setosa 35 4.9 3.1 1.5 .2 setosa 36 5.0 3.2 1.2 .2 setosa 37 5.5 3.5 1.3 .2 setosa 38 4.9 3.6 1.4 .1 setosa 39 4.4 3.0 1.3 .2 setosa 40 5.1 3.4 1.5 .2 setosa 41 5.0 3.5 1.3 .3 setosa 42 4.5 2.3 1.3 .3 setosa 43 4.4 3.2 1.3 .2 setosa 44 5.0 3.5 1.6 .6 setosa 45 5.1 3.8 1.9 .4 setosa 46 4.8 3.0 1.4 .3 setosa 47 5.1 3.8 1.6 .2 setosa 48 4.6 3.2 1.4 .2 setosa 49 5.3 3.7 1.5 .2 setosa 50 5.0 3.3 1.4 .2 setosa 51 7.0 3.2 4.7 1.4 versicolor 52 6.4 3.2 4.5 1.5 versicolor 53 6.9 3.1 4.9 1.5 versicolor 54 5.5 2.3 4.0 1.3 versicolor 55 6.5 2.8 4.6 1.5 versicolor 56 5.7 2.8 4.5 1.3 versicolor 57 6.3 3.3 4.7 1.6 versicolor 58 4.9 2.4 3.3 1.0 versicolor 59 6.6 2.9 4.6 1.3 versicolor 60 5.2 2.7 3.9 1.4 versicolor 61 5.0 2.0 3.5 1.0 versicolor 62 5.9 3.0 4.2 1.5 versicolor 63 6.0 2.2 4.0 1.0 versicolor 64 6.1 2.9 4.7 1.4 versicolor 65 5.6 2.9 3.6 1.3 versicolor 66 6.7 3.1 4.4 1.4 versicolor 67 5.6 3.0 4.5 1.5 versicolor 68 5.8 2.7 4.1 1.0 versicolor 69 6.2 2.2 4.5 1.5 versicolor 70 5.6 2.5 3.9 1.1 versicolor 71 5.9 3.2 4.8 1.8 versicolor 72 6.1 2.8 4.0 1.3 versicolor 73 6.3 2.5 4.9 1.5 versicolor 74 6.1 2.8 4.7 1.2 versicolor 75 6.4 2.9 4.3 1.3 versicolor 76 6.6 3.0 4.4 1.4 versicolor 77 6.8 2.8 4.8 1.4 versicolor 78 6.7 3.0 5.0 1.7 versicolor 79 6.0 2.9 4.5 1.5 versicolor 80 5.7 2.6 3.5 1.0 versicolor 81 5.5 2.4 3.8 1.1 versicolor 82 5.5 2.4 3.7 1.0 versicolor 83 5.8 2.7 3.9 1.2 versicolor 84 6.0 2.7 5.1 1.6 versicolor 85 5.4 3.0 4.5 1.5 versicolor 86 6.0 3.4 4.5 1.6 versicolor 87 6.7 3.1 4.7 1.5 versicolor 88 6.3 2.3 4.4 1.3 versicolor 89 5.6 3.0 4.1 1.3 versicolor 90 5.5 2.5 4.0 1.3 versicolor 91 5.5 2.6 4.4 1.2 versicolor 92 6.1 3.0 4.6 1.4 versicolor 93 5.8 2.6 4.0 1.2 versicolor 94 5.0 2.3 3.3 1.0 versicolor 95 5.6 2.7 4.2 1.3 versicolor 96 5.7 3.0 4.2 1.2 versicolor 97 5.7 2.9 4.2 1.3 versicolor 98 6.2 2.9 4.3 1.3 versicolor 99 5.1 2.5 3.0 1.1 versicolor 100 5.7 2.8 4.1 1.3 versicolor 101 6.3 3.3 6.0 2.5 virginica 102 5.8 2.7 5.1 1.9 virginica 103 7.1 3.0 5.9 2.1 virginica 104 6.3 2.9 5.6 1.8 virginica 105 6.5 3.0 5.8 2.2 virginica 106 7.6 3.0 6.6 2.1 virginica 107 4.9 2.5 4.5 1.7 virginica 108 7.3 2.9 6.3 1.8 virginica 109 6.7 2.5 5.8 1.8 virginica 110 7.2 3.6 6.1 2.5 virginica 111 6.5 3.2 5.1 2.0 virginica 112 6.4 2.7 5.3 1.9 virginica 113 6.8 3.0 5.5 2.1 virginica 114 5.7 2.5 5.0 2.0 virginica 115 5.8 2.8 5.1 2.4 virginica 116 6.4 3.2 5.3 2.3 virginica 117 6.5 3.0 5.5 1.8 virginica 118 7.7 3.8 6.7 2.2 virginica 119 7.7 2.6 6.9 2.3 virginica 120 6.0 2.2 5.0 1.5 virginica 121 6.9 3.2 5.7 2.3 virginica 122 5.6 2.8 4.9 2.0 virginica 123 7.7 2.8 6.7 2.0 virginica 124 6.3 2.7 4.9 1.8 virginica 125 6.7 3.3 5.7 2.1 virginica 126 7.2 3.2 6.0 1.8 virginica 127 6.2 2.8 4.8 1.8 virginica 128 6.1 3.0 4.9 1.8 virginica 129 6.4 2.8 5.6 2.1 virginica 130 7.2 3.0 5.8 1.6 virginica 131 7.4 2.8 6.1 1.9 virginica 132 7.9 3.8 6.4 2.0 virginica 133 6.4 2.8 5.6 2.2 virginica 134 6.3 2.8 5.1 1.5 virginica 135 6.1 2.6 5.6 1.4 virginica 136 7.7 3.0 6.1 2.3 virginica 137 6.3 3.4 5.6 2.4 virginica 138 6.4 3.1 5.5 1.8 virginica 139 6.0 3.0 4.8 1.8 virginica 140 6.9 3.1 5.4 2.1 virginica 141 6.7 3.1 5.6 2.4 virginica 142 6.9 3.1 5.1 2.3 virginica 143 5.8 2.7 5.1 1.9 virginica 144 6.8 3.2 5.9 2.3 virginica 145 6.7 3.3 5.7 2.5 virginica 146 6.7 3.0 5.2 2.3 virginica 147 6.3 2.5 5.0 1.9 virginica 148 6.5 3.0 5.2 2.0 virginica 149 6.2 3.4 5.4 2.3 virginica 150 5.9 3.0 5.1 1.8 virginica Example PCA biplot using the iris data set in R (code below):
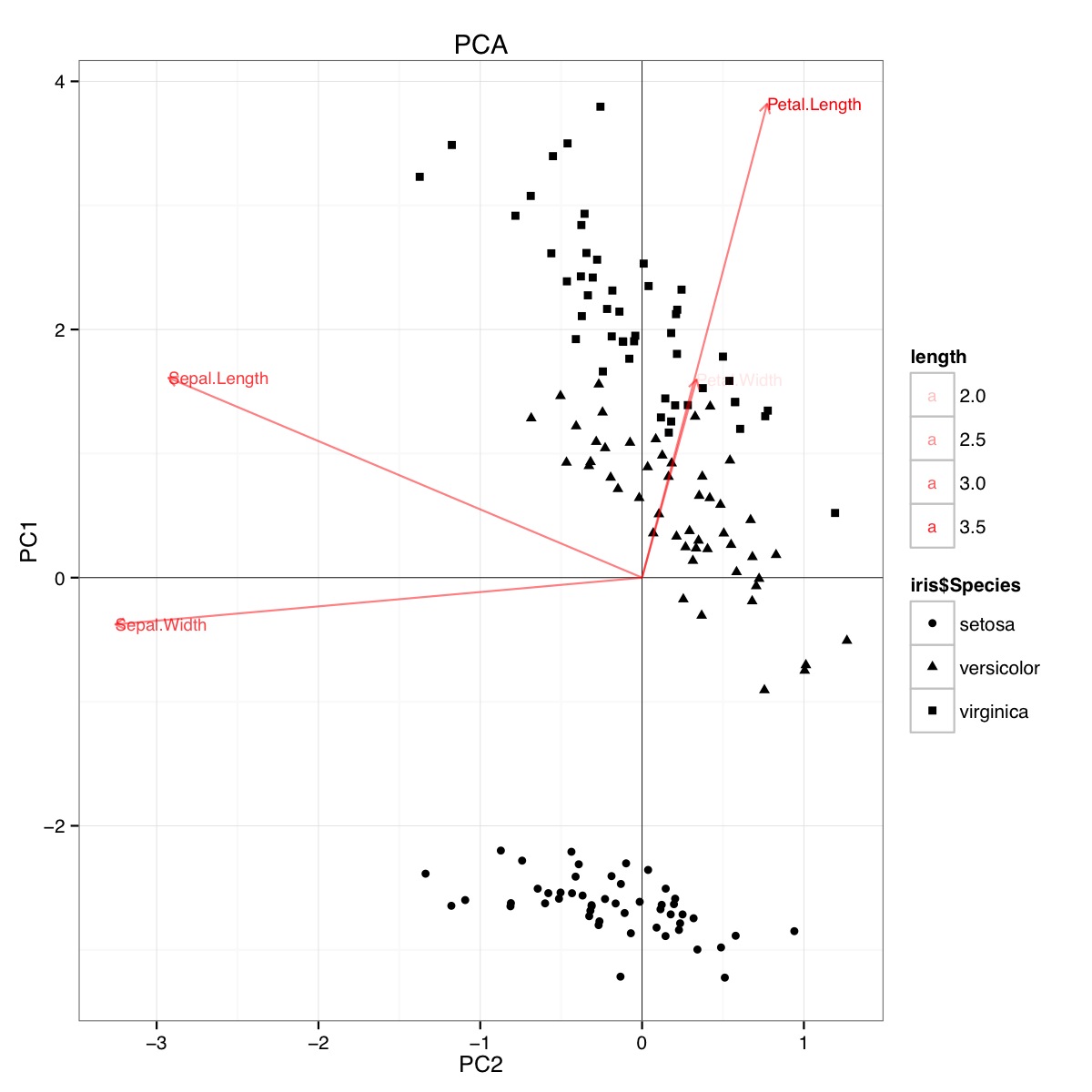
This figure indicates that Petal length and Petal width are important in determining PC1 score and in discriminating between Species groups. setosa has smaller petals and wider sepals.
Apparently, similar conclusions can be drawn from plotting linear discriminant analysis results, though I am not certain what the LDA plot presents, hence the question. The axis are the two first linear discriminants (LD1 99% and LD2 1% of trace). The coordinates of the red vectors are "Coefficients of linear discriminants" also described as "scaling" (lda.fit$scaling: a matrix which transforms observations to discriminant functions, normalized so that within groups covariance matrix is spherical). "scaling" is calculated as diag(1/f1, , p) and f1 is sqrt(diag(var(x - group.means[g, ]))). Data can be projected onto the linear discriminants (using predict.lda) (code below, as demonstrated https://stackoverflow.com/a/17240647/742447). The data and the predictor variables are plotted together so that which species are defined by an increase in which predictor variables can be seen (as is done for usual PCA biplots and the above PCA biplot).:
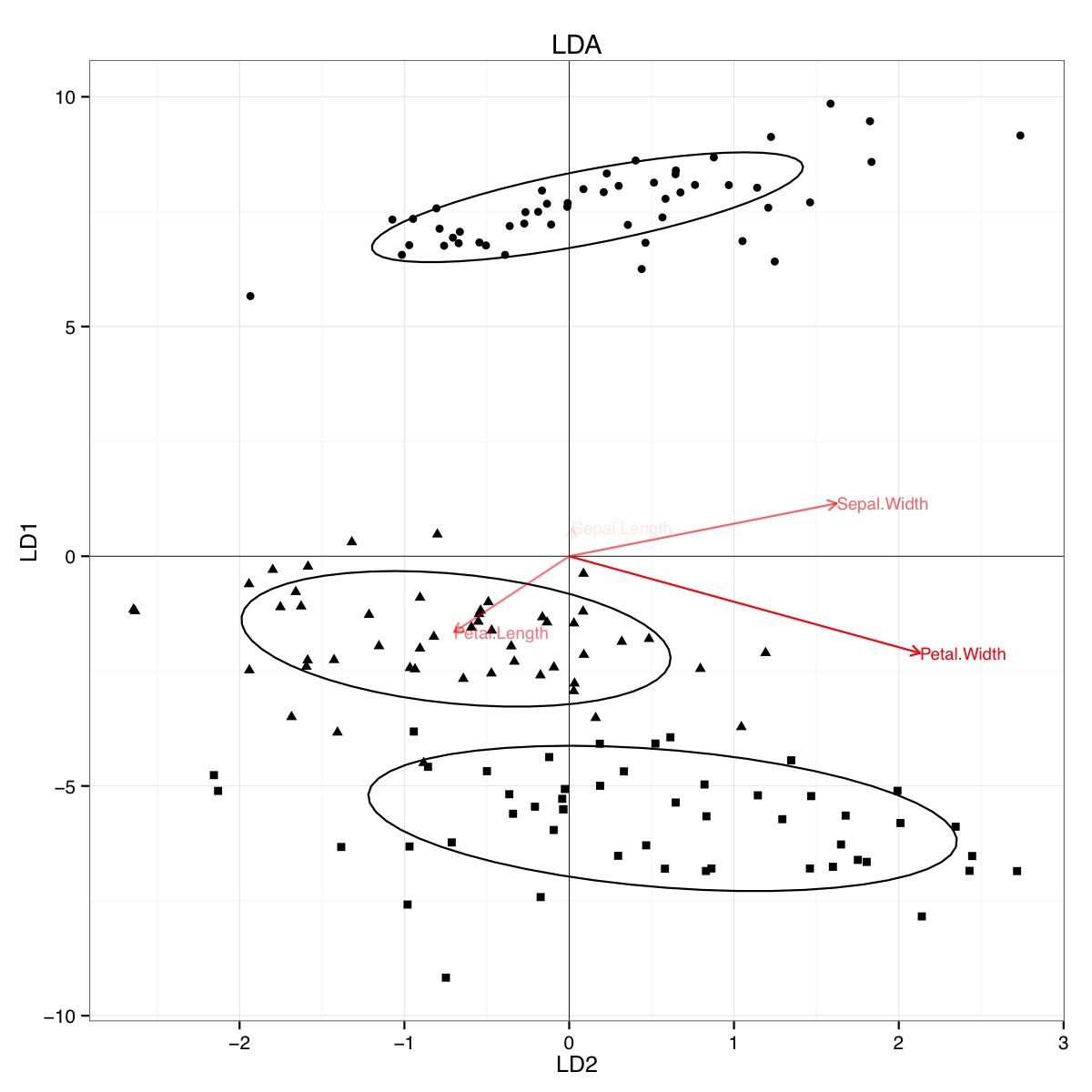
From this plot, Sepal width, Petal Width and Petal Length all contribute to a similar level to LD1. As expected, setosa appears to smaller petals and wider sepals.
There is no built-in way to plot such biplots from LDA in R and few discussions of this online, which makes me wary of this approach.
Does this LDA plot (see code below) provide a statistically valid interpretation of predictor variable scaling scores ?
Code for PCA:
require(grid) iris.pca <- prcomp(iris[,-5]) PC <- iris.pca x="PC1" y="PC2" PCdata <- data.frame(obsnames=iris[,5], PC$x) datapc <- data.frame(varnames=rownames(PC$rotation), PC$rotation) mult <- min( (max(PCdata[,y]) - min(PCdata[,y])/(max(datapc[,y])-min(datapc[,y]))), (max(PCdata[,x]) - min(PCdata[,x])/(max(datapc[,x])-min(datapc[,x]))) ) datapc <- transform(datapc, v1 = 1.6 * mult * (get(x)), v2 = 1.6 * mult * (get(y)) ) datapc$length <- with(datapc, sqrt(v1^2+v2^2)) datapc <- datapc[order(-datapc$length),] p <- qplot(data=data.frame(iris.pca$x), main="PCA", x=PC1, y=PC2, shape=iris$Species) #p <- p + stat_ellipse(aes(group=iris$Species)) p <- p + geom_hline(aes(0), size=.2) + geom_vline(aes(0), size=.2) p <- p + geom_text(data=datapc, aes(x=v1, y=v2, label=varnames, shape=NULL, linetype=NULL, alpha=length), size = 3, vjust=0.5, hjust=0, color="red") p <- p + geom_segment(data=datapc, aes(x=0, y=0, xend=v1, yend=v2, shape=NULL, linetype=NULL, alpha=length), arrow=arrow(length=unit(0.2,"cm")), alpha=0.5, color="red") p <- p + coord_flip() print(p) Code for LDA
#Perform LDA analysis iris.lda <- lda(as.factor(Species)~., data=iris) #Project data on linear discriminants iris.lda.values <- predict(iris.lda, iris[,-5]) #Extract scaling for each predictor and data.lda <- data.frame(varnames=rownames(coef(iris.lda)), coef(iris.lda)) #coef(iris.lda) is equivalent to iris.lda$scaling data.lda$length <- with(data.lda, sqrt(LD1^2+LD2^2)) scale.para <- 0.75 #Plot the results p <- qplot(data=data.frame(iris.lda.values$x), main="LDA", x=LD1, y=LD2, shape=iris$Species)#+stat_ellipse() p <- p + geom_hline(aes(0), size=.2) + geom_vline(aes(0), size=.2) p <- p + theme(legend.position="none") p <- p + geom_text(data=data.lda, aes(x=LD1*scale.para, y=LD2*scale.para, label=varnames, shape=NULL, linetype=NULL, alpha=length), size = 3, vjust=0.5, hjust=0, color="red") p <- p + geom_segment(data=data.lda, aes(x=0, y=0, xend=LD1*scale.para, yend=LD2*scale.para, shape=NULL, linetype=NULL, alpha=length), arrow=arrow(length=unit(0.2,"cm")), color="red") p <- p + coord_flip() print(p) The results of the LDA are as follows
lda(as.factor(Species) ~ ., data = iris) Prior probabilities of groups: setosa versicolor virginica 0.3333333 0.3333333 0.3333333 Group means: Sepal.Length Sepal.Width Petal.Length Petal.Width setosa 5.006 3.428 1.462 0.246 versicolor 5.936 2.770 4.260 1.326 virginica 6.588 2.974 5.552 2.026 Coefficients of linear discriminants: LD1 LD2 Sepal.Length 0.8293776 0.02410215 Sepal.Width 1.5344731 2.16452123 Petal.Length -2.2012117 -0.93192121 Petal.Width -2.8104603 2.83918785 Proportion of trace: LD1 LD2 0.9912 0.0088 
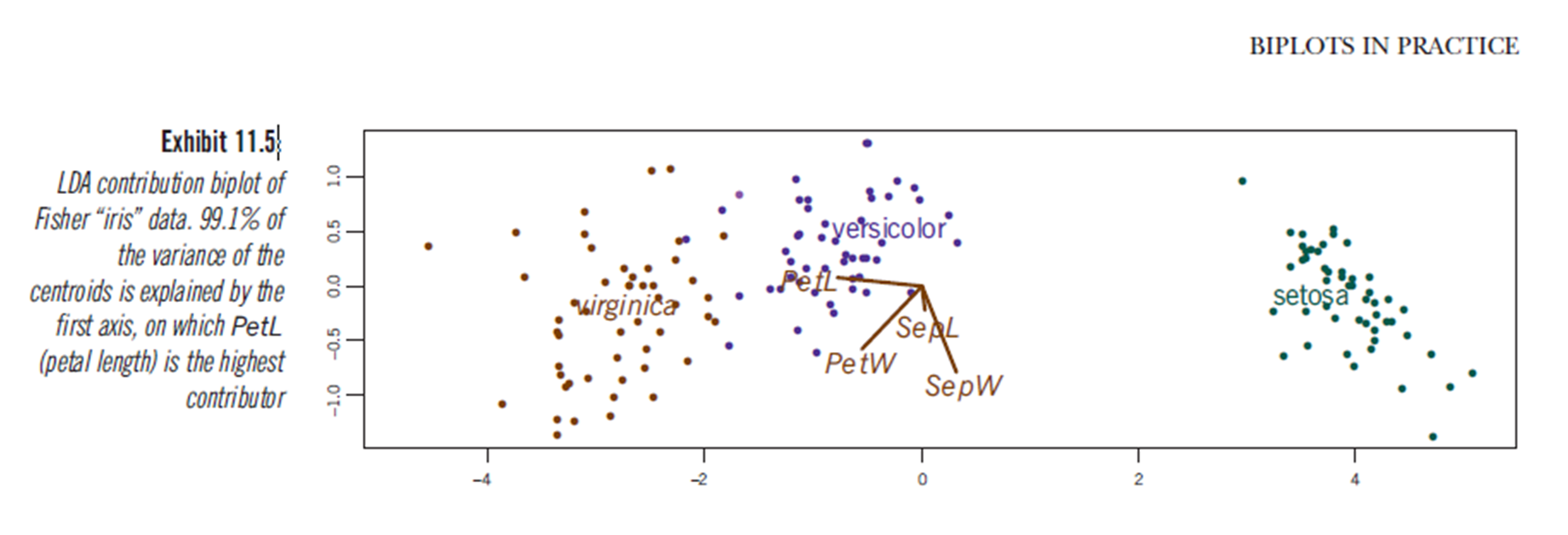
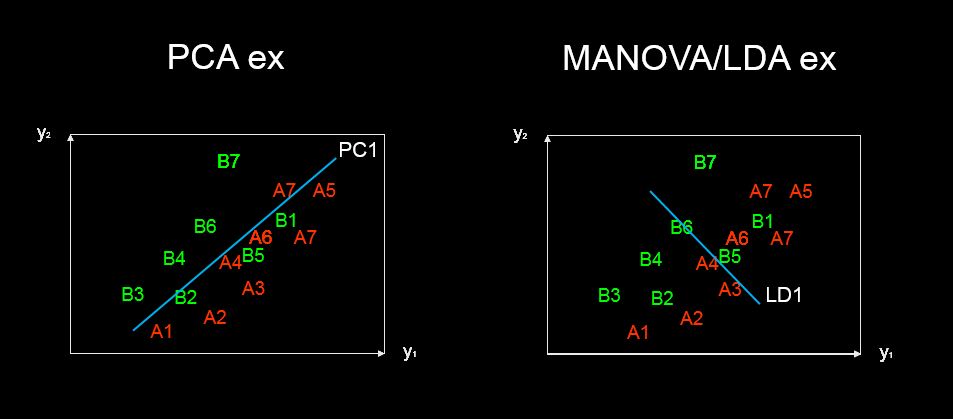
discriminant predictor variable scaling scores? - the term seems to me not common and strange. $\endgroup$predictor variable scaling scores. Maybe "discriminant scores"? Anyway, I added an answer which might be of your interest. $\endgroup$